ABSTRACT
Secondary metabolites in horticultural crops encompass a wide array of bioactive compounds that are crucial for plant defense and contribute to flavor, color, aroma, and human health benefits. Increasing demand for plant derived natural products in the pharmaceutical, nutraceutical, cosmetic, and food industries has intensified research aimed at optimizing their production and extraction. Conventional cultivation methods, which depend on field-grown plants, are constrained by slow growth, seasonal fluctuations, and low metabolite yields. In contrast, biotechnological approaches provide efficient alternatives for enhancing secondary metabolite accumulation. Metabolic engineering enables targeted modification of biosynthetic pathways through gene overexpression, gene silencing, and the introduction of heterologous genes. Elicitation strategies stimulate defense-related metabolic pathways in response to biotic and abiotic factors. This review critically examines recent progress in both approaches for horticultural crops and explores integrated strategies that combine metabolic pathway engineering with elicitor-mediated induction. The review also highlights the application of omics technologies and CRISPR/Cas-based genome editing. Future directions underscore the potential of synthetic biology and systems-level modeling to achieve predictable and sustainable engineering of secondary metabolite biosynthesis in horticultural crops.
Keywords: Secondary Metabolites; Metabolic Engineering; Elicitation; Omics Technologies; CRISPR/Cas
INTRODUCTION
Horticultural crops, including fruits, vegetables, spices, medicinal, and ornamental species, play a vital role in human health, cultural traditions, and the global economy [1]. In addition to primary metabolic processes that support plant growth and reproduction, these crops synthesize a diverse range of secondary metabolites, such as phenolics, terpenoids, alkaloids, flavonoids, and glycosides. These compounds determine organoleptic properties, enhance environmental adaptability, and confer significant health benefits [2, 3]. Furthermore, these bioactive molecules contribute aroma, flavor, pigmentation, and nutritional quality, and serve as essential raw materials for the nutraceutical, pharmaceutical, and cosmetic industries [4].
The biosynthesis of these compounds is complex, involving multi-step enzymatic reactions within pathways such as the phenylpropanoid, mevalonate (MVA), and Methyl Erythritol Phosphate (MEP) networks. Regulation occurs at transcriptional, translational, and post-translational levels, with key enzymes subject to feedback inhibition and substrate competition [5]. As a result, natural yields of many high-value metabolites are low, and their accumulation is strongly influenced by environmental factors, including light quality, temperature, salinity, and pathogen pressure, which modulate gene expression and enzymatic activity [6, 7]. Traditional plant breeding and agronomic practices often fail to improve secondary metabolite content because these traits are polygenic, quantitatively inherited, and highly responsive to environmental variation [8].
In the past two decades, metabolic engineering has become a key strategy for manipulating metabolic fluxes within biosynthetic pathways. This approach includes overexpressing or silencing rate-limiting enzymes, introducing heterologous genes, and modifying transcription factors such as MYB, bHLH, and WRKY, which regulate metabolite biosynthesis [1]. In parallel, elicitation strategies utilize biotic (such as chitosan and yeast extract), abiotic (such as UV light and temperature shifts), or chemical (such as methyl jasmonate and salicylic acid) stimuli to activate endogenous defense signaling. These interventions enhance the accumulation of secondary metabolites without permanent genetic modification [2, 8].
The integration of omics technologies, including genomics, transcriptomics, proteomics, and metabolomics, has significantly advanced the understanding of complex regulatory networks underlying secondary metabolism [3]. Systems biology and machine learning models are increasingly used to predict pathway bottlenecks, simulate metabolite flux, and inform rational design for yield optimization [9]. When combined with CRISPR-based genome editing, synthetic promoters, and automated bioreactor systems, these innovations facilitate dynamic and sustainable production of high-value metabolites in controlled environments [5].
Despite significant advances, a comprehensive framework that integrates metabolic engineering and elicitation for enhancing secondary metabolites in horticultural crops is still lacking. Most research examines these strategies separately, without providing a mechanistic or translational synthesis that connects genetic regulation, elicitor signaling, and yield scalability [10]. This review therefore addresses the following central question:
How can metabolic engineering and elicitation be strategically integrated to achieve predictable, scalable, and sustainable enhancement of secondary metabolites in horticultural crops?
To address this question, the review systematically examines metabolic engineering targets and regulatory networks, elicitor types and signal transduction pathways, quantitative case studies across key horticultural species, and translational considerations for commercial application. The central hypothesis is that a synergistic combination of intrinsic (genetic) and extrinsic (elicitor-mediated) controls yields greater and more stable metabolite production than either strategy alone.
This review focuses specifically on horticultural crops cultivated for food, flavor, fragrance, and pharmaceutical applications, integrating metabolic engineering and elicitation strategies within this context. Particular emphasis is given to species such as Capsicum annuum, Ocimum basilicum, Mentha piperita, and Solanum lycopersicum, where combined approaches have demonstrated measurable success.
Overview of Secondary Metabolites in Horticultural Crops
Secondary metabolites in horticultural crops comprise a chemically diverse group of bioactive compounds, primarily classified as terpenoids, phenolics, and nitrogen-containing metabolites, including alkaloids and glycosides. These metabolites are synthesized through distinct but interconnected biosynthetic pathways: the phenylpropanoid pathway, which produces phenolics and flavonoids, and the isoprenoid pathways, specifically the mevalonate (MVA) and Methyl Erythritol Phosphate (MEP) routes, which generate terpenoids [11]. The synthesis and accumulation of these compounds are regulated by developmental stage, tissue specificity, and environmental factors. Major transcription factor families, such as MYB, bHLH, WRKY, and AP2/ERF, control the expression of structural genes in response to stress and signaling networks [12]. In horticultural species, secondary metabolites are key determinants of organoleptic and functional quality. For example, terpenoids contribute to aroma and essential oil profiles in Ocimum basilicum and Mentha piperita [13,14]; phenolics, including anthocyanins and flavonoids, influence fruit and flower pigmentation in Vitis vinifera and Solanum lycopersicum [15]; and alkaloids such as capsaicinoids in Capsicum annuum and limonoids in Citrus sinensis provide pharmacological and nutraceutical benefits [16,17]. Due to environmental variability and low yields in field-grown plants, in vitro culture systems—including callus, cell suspensions, hairy roots, and microrhizomes—have been developed as controlled alternatives for producing high-value metabolites independent of seasonal constraints [18,19]. In Zingiber officinale (ginger), for example, in vitro elicitation strategies and optimized culture conditions have increased the accumulation of gingerol and shogaol [18]. However, production bottlenecks such as feedback inhibition and pathway competition continue to limit large-scale metabolite yields. To address these challenges, integrative approaches that combine transcriptional and metabolic engineering, elicitor-mediated induction, and pathway redesign are increasingly utilized to achieve sustainable and commercially viable biosynthesis of secondary metabolites in horticultural crops [11,20].
MAIN REVIEW CONTENT
Metabolic Engineering Strategies
Metabolic engineering entails the targeted modification of metabolic networks through genetic and computational methodologies to redirect metabolic flux toward specific metabolites. Principal strategies encompass gene overexpression, RNA interference (RNAi), transcription factor modulation, and pathway reconstruction utilizing heterologous systems.
Pathway Gene Manipulation
Manipulation of key pathway genes is fundamental to metabolic engineering efforts aimed at increasing secondary metabolite production in horticultural crops. Overexpressing rate-limiting enzymes enhances metabolic flux and promotes the accumulation of target compounds. For example, in tomato ( Solanum lycopersicum), co-expression of chalcone synthase (CHS) and Flavonol Synthase (FLS) significantly increased flavonol and anthocyanin content, illustrating the effectiveness of multigene overexpression in augmenting phenylpropanoid metabolism [21]. In Mentha species, modulation of enzymes such as limonene synthase and menthofuran synthase alters terpene profiles, thereby improving aromatic quality and oil yield [22]. In addition to overexpression, pathway manipulation includes silencing or editing of competitive branches that divert metabolic precursors. RNA interference (RNAi)-mediated suppression of Phenylalanine Ammonia-Lyase (PAL) has redirected flux toward anthocyanin and flavonoid biosynthesis in grape, Epimedium, and other plant systems [23]. Recent advances in CRISPR/Cas-based editing enable precise regulation of enzyme-encoding genes, facilitating the rebalancing of pathway fluxes and improving both yield and metabolite specificity in targeted horticultural crops [24,25]. Overall, pathway gene engineering, which integrates enzyme overexpression, RNAi silencing, and genome editing, establishes a rational framework for predictable and sustainable tailoring of biosynthetic output.
Transcription Factor Engineering
Transcription factors (TFs) serve as central regulators of plant secondary metabolism by coordinating the expression of biosynthetic pathway genes. Major TF families, including MYB, bHLH, and WRKY proteins, are essential for activating or repressing key steps in phenylpropanoid, terpenoid, and alkaloid biosynthesis. For example, overexpression of AtMYB12 in tomato ( Solanum lycopersicum) significantly increased flavonol accumulation and altered gene expression within the phenylpropanoid pathway [26,27]. Similarly, SmMYB1, a methyl jasmonate–responsive transcription factor from Salvia miltiorrhiza, promotes phenolic acid and tanshinone biosynthesis by activating CYP98A14 and related downstream enzymes [28]. Recent reviews indicate that MYB transcription factors function as broad-spectrum regulators, influencing plant growth, stress responses, and secondary metabolism [29,30]. Other families, such as bHLH and WRKY, interact with MYB proteins to form regulatory complexes that fine-tune metabolite output in response to environmental or hormonal signals [24,25]. Advances in genome editing and synthetic biology now enable precise modulation of these transcriptional regulators to enhance target metabolite production without negatively affecting plant fitness [31,32]. Engineering TFs, including MYB–bHLH–WD40 complexes, provides a means to simultaneously activate multiple biosynthetic genes, resulting in increased yields of flavonoids, terpenes, and alkaloids in horticultural crops. Thus, transcription factor engineering is a promising area in metabolic engineering, allowing for predictable, heritable, and multi-gene control of secondary metabolite biosynthesis in economically important plants.
RNA interference, CRISPR/Cas and Synthetic Biology Approaches
RNA interference (RNAi) and CRISPR/Cas genome-editing systems facilitate targeted suppression of undesirable or competing pathways, thereby enabling metabolic reprogramming in plants [33,34]. For example, manipulation of the cytochrome P450 gene CYP76AD1, which catalyzes the initial tyrosine-hydroxylation step in betalain pigment biosynthesis in Beta vulgaris, redirects carbon flux toward pigment or other secondary metabolite pathways [35]. Similarly, CRISPR-mediated mutagenesis of acyltransferase genes such as Pun1 in Capsicum annuum has altered capsaicinoid synthesis, demonstrating the utility of genome editing for trait improvement in horticultural crops [36]. In addition to gene knockout, synthetic biology approaches are increasingly employed, assembling modular multi-gene pathways in plant or microbial systems to redirect metabolite flow and enhance accumulation of target compounds. Recent reviews underscore the role of systems biology and synthetic biology techniques in elucidating and engineering plant natural product pathways [37,38]. Collectively, these converging technologies RNAi, CRISPR/Cas, and synthetic biology are rapidly increasing the precision and scope of metabolic improvement in non-model horticultural crops.
Omics Integration and Computational Modelling
The integration of multi-omics datasets, including genomics, transcriptomics, proteomics, and metabolomics, enables a comprehensive systems-level understanding of plant biosynthetic networks. This approach facilitates the identification of key regulatory nodes and metabolic pathways, supporting targeted interventions in metabolic engineering. Recent advances in machine learning have improved the prediction of genes involved in specialized metabolite biosynthesis. For example, applied the automated machine learning framework AutoGluon-Tabular to integrate multi-omics data from Arabidopsis, successfully predicting genes encoding enzymes for terpenoid, alkaloid, and phenolic biosynthesis [39]. This demonstrates the utility of machine learning in elucidating complex biosynthetic pathways across diverse plant species.
In addition to data-driven approaches, computational modeling techniques such as Flux Balance Analysis (FBA) are essential for simulating metabolic flux distributions in reconstructed genome-scale models. These models help identify metabolic bottlenecks and potential targets for genetic modification. Recent reviews have emphasized the integration of multi-omics data into metabolic models, highlighting both the challenges and opportunities associated with applying network flux analysis to plant systems [40,41].
Moreover, integrating multi-omics data with computational tools has enabled the development of unsupervised workflows, such as MEANtools, which connect transcriptomic and metabolomic data to predict metabolic pathways without prior knowledge [42]. This strategy supports the discovery of novel biosynthetic routes and advances the understanding of plant metabolic networks.
The convergence of multi-omics integration and computational modeling accelerates the rational design of metabolic pathways, facilitating the optimization of metabolite production in horticultural crops. These integrative strategies are essential for advancing plant metabolic engineering and provide new opportunities for crop improvement and sustainable agriculture.
Subcellular Targeting and Transporter Engineering
Improving the storage and transport of specialized metabolites within plant cells is essential for increasing yield and stability. Engineering transporter proteins, such as ATP-Binding Cassette (ABC) transporters, enables the sequestration of compounds like alkaloids and phenolics into vacuoles, which stabilizes product accumulation and reduces cytotoxicity [43]. For example, ABC transporters are involved in the vacuolar sequestration of various secondary metabolites, including alkaloids and phenolics, thereby contributing to their detoxification and storage within plant cells [44].
The Multidrug and Toxin Extrusion (MATE) transporter family also plays a significant role in the vacuolar sequestration of flavonoid glycosides. Functional characterization of MATE2 from Medicago truncatula demonstrated its involvement in transporting anthocyanins and other flavonoid glycosides into vacuoles, thereby increasing their accumulation and stability [45].
The Nitrate and Peptide Transporter (NPF) family has also emerged as a key component in the transport of various metabolites, including specialized compounds. Recent studies have shown that NPF transporters contribute to the long-distance transport and homeostasis of iron, independent of their nitrate transport function, indicating a broader role in plant specialized metabolism [46].
Engineering transporter families such as ABC, MATE, and NPF presents a promising strategy for increasing the accumulation and stability of valuable secondary metabolites in plants. Manipulating the expression and activity of these transporters can optimize intracellular localization and storage of metabolites, thereby improving yield and supporting the development of crops with enhanced nutritional and pharmaceutical value.
Elicitation Strategies
Elicitors are external stimuli, either biotic or abiotic, that simulate stress responses and thereby enhance the synthesis of secondary metabolites. Elicitation represents an efficient and cost-effective strategy that is compatible with in vitro culture systems.
Chemical Elicitors
Chemical elicitors, including Methyl Jasmonate (MeJA), Salicylic Acid (SA), and Abscisic Acid (ABA), play critical roles in activating plant defense-signaling pathways that simulate biotic and abiotic stress conditions, thereby enhancing the biosynthesis of specialized metabolites. MeJA, in particular, is recognized as a highly potent elicitor, significantly increasing the production of terpenoids, flavonoids, and other secondary metabolites in a wide range of plant species. For instance, treatment of adventitious root cultures of Panax ginseng with MeJA led to a substantial increase in total saponin (ginsenoside) content and up-regulation of triterpenoid biosynthesis genes [47]. In Vitis vinifera (grapevine), exogenous MeJA application enhanced resveratrol and stilbene accumulation in berry clusters by activating jasmonate-responsive transcription factors and downstream biosynthetic genes [48]. In Ocimum basilicum (basil), MeJA treatment increased levels of rosmarinic acid and eugenol, which are key phenolic and volatile compounds with antioxidant and antimicrobial properties, through up-regulation of phenylpropanoid pathway genes [49]. Collectively, these findings demonstrate the effectiveness of chemical elicitors, particularly MeJA, in stimulating metabolic pathways and improving the yield and quality of valuable phytochemicals in horticultural crops.
Biotic Elicitors
Biotic elicitors, which are derived from living organisms or their cell-wall components, play a critical role in enhancing secondary metabolite production by simulating pathogen attack and activating the plant’s innate defense mechanisms. These elicitors include microbial extracts (from bacteria, fungi, and yeast), polysaccharides such as chitosan, pectin, and glucans, as well as cell-wall fragments like chitin. Notably, chitosan, which is obtained from fungal or crustacean sources, functions as a Pattern-Associated Molecular Pattern (PAMP) and triggers bursts of Reactive Oxygen Species (ROS), calcium influx, and activation of defense-related enzymes including Phenylalanine Ammonia-Lyase (PAL), peroxidases, and polyphenol oxidases [50]. Studies have shown that chitosan application in in vitro culture systems significantly increases phenolic and flavonoid content [50]. For example, suspension cultures of Vitis vinifera treated with yeast extract exhibited substantial increases in phenolic accumulation due to the overexpression of jasmonate-responsive genes [51]. Reviews of biotic elicitation mechanisms indicate that bacterial, fungal, and algal elicitors can increase the biosynthesis of valuable compounds by two- to more than ten-fold, depending on culture conditions and plant species [51]. Although these findings confirm the broad applicability of biotic elicitors in horticultural and medicinal plants, challenges persist in standardizing elicitor type, dosage, timing, and culture systems to optimize metabolite yields [52]. In summary, biotic elicitors offer a high-potential and cost-effective strategy for enhancing secondary metabolite production in horticultural biotechnology.
Abiotic Elicitors
Physical stresses, such as Ultraviolet (UV) irradiation, temperature fluctuations, salinity, and other abiotic factors, can enhance phenolic and carotenoid biosynthesis by activating oxidative signaling and photoreceptor-mediated pathways. For instance, UV-B radiation applied to red cabbage sprouts increased Total Phenolic Content (TPC) and Total Flavonoid Content (TFC) by approximately 35% and 30%, respectively, and also enhanced carotenoid accumulation at specific UV-B doses [53]. Reviews suggest that these abiotic stressors function by inducing Reactive Oxygen Species (ROS), activating phenylpropanoid pathway enzymes such as Phenylalanine Ammonia-Lyase (PAL), and modulating light signaling and photomorphogenic regulators [54]. Additionally, controlled light treatments in leafy vegetables have shown that varying light spectra can modulate carotenoid biosynthesis through photoreceptor and ROS-mediated regulatory networks [55]. Therefore, abiotic elicitors provide an effective strategy for increasing secondary metabolite accumulation in horticultural crops by leveraging stress-induced signaling rather than relying on direct genetic modification.
Nanoparticle-Based Elicitation
Engineered nanoparticles (NPs), including metallic (AgNPs, AuNPs), metal-oxide (ZnO, TiO₂), and carbon-based nanomaterials, have emerged as innovative elicitors of secondary metabolism in plants by penetrating tissues, modulating redox balance, and initiating oxidative stress signaling cascades [56]. For example, application of AgNPs at sub-toxic concentrations has been shown to generate reactive oxygen species (ROS) and activate MAPK pathways, which up-regulate Phenylalanine Ammonia-Lyase (PAL) and other biosynthetic enzymes, resulting in increased phenolic content in Catharanthus roseus cultures [57]. Additionally, AgNP treatments have led to elevated levels of vincristine and vinblastine, compounds with significant therapeutic potential in cancer treatment, and have demonstrated synergistic effects when combined with other elicitors such as methyl jasmonate (MeJA) [58]. More broadly, several reviews indicate that NPs can enhance the accumulation of flavonoids and alkaloids in both in vitro and in vivo systems by acting as abiotic elicitors that mimic stress responses [59]. As a result, nano-elicitation integrates physical properties (such as size and surface charge) and chemical signaling (including ion release and ROS induction), positioning it as a leading technology for increasing high-value phytochemical production in horticultural crops.
Signal Transduction Pathways
Elicitor perception in plants typically begins when Pattern Recognition Receptors (PRRs) and Receptor-Like Kinases (RLKs) detect external cues such as pathogen-associated molecular patterns (PAMPs) or Damage-Associated Molecular Patterns (DAMPs), which initiate downstream signaling cascades [60,61]. Early responses include calcium ion (Ca²⁺) influx, generation of reactive oxygen species (ROS), and activation of Mitogen-Activated Protein Kinase (MAPK) and Calcium-Dependent Protein Kinase (CDPK) pathways, which function as central signaling hubs in elicitor-response networks [62]. These primary signals integrate with phytohormone pathways, particularly Jasmonic Acid (JA), Ethylene (ET), and Salicylic Acid (SA), to regulate transcriptional networks that control secondary metabolism [63,64]. For example, JA signaling coordinates the biosynthesis of defense-related secondary metabolites, such as alkaloids and flavonoids, through MAPK-mediated activation of downstream genes [65]. Similarly, the interaction between SA and ROS pathways contributes to Systemic Acquired Resistance (SAR) and modulates phenolic and terpenoid biosynthesis [66]. Collectively, these interconnected signaling systems enable elicitor perception at the cell membrane to drive metabolic reprogramming and enhance the accumulation of valuable secondary metabolites in horticultural crops.
|
Type of Elicitor |
Example(s) |
Mode of Action |
Target Metabolites |
Crops |
Observed Effect |
Reference |
|
Chemical |
Methyl jasmonate (MeJA) |
Activates jasmonate-signaling and defense-related transcriptional cascades |
Polyphenols, |
Oryza sativa |
Enhanced production of resveratrol and piceid |
[67] |
|
Alkaloids, |
Tinospora cordifolia |
5.57-fold higher than berberine content |
[68] |
|||
|
Flavonoids, |
Vernonia anthelmintica |
2.2-fold higher accumulation of rhamnetin |
[69] |
|||
|
Terpinoids |
Citrullus colocynthis |
Highest accumulation of cucurbitacin E |
[70] |
|||
|
Biotic |
Chitosan, yeast extract |
Mimics pathogen attack and triggers ROS-mediated phenylpropanoid pathway |
Phenolics, alkaloids |
Dracocephalum kotschyi |
chitosan spray had a significant influence on the principle essential oil components, such as p-cymene and thymol. |
[71] |
|
Trigonella foenum-graecum |
Co-60 gamma-irradiated chitosan significantly boosted the total alkaloid content, seed yield, and trigonelline constituent. |
|||||
|
Perovskia abrotanoides |
Yeast extract increased Cryptotanshinone and tanshinone IIA |
[73] |
||||
|
Hypericum perforatum and Vitis vinifera |
Chitin increased Phenylpropanoid and Naphtodianthrone, trans-Resveratrol and Viniferins, |
[74] |
||||
|
Chitin and Pectin increased Hypericin and Pseudohypericin, |
[75] |
|||||
|
Taverniera cuneifolia |
Rhizobium leguminosarum increased Glycyrrhizic acid |
[76] |
||||
|
Abiotic |
UV light, salinity, temperature, drought |
Induces oxidative signaling and enhances antioxidant enzyme activity |
Flavonoids, carotenoids, phenolics |
Centella asiatica |
UV-B and High PARS increased saponins |
[77] |
|
Astragalus compactus |
High temperature increased phenolics |
[78] |
||||
|
Plantago ovata |
NaCl increased Flavonoids |
[79] |
||||
|
Brassica oleracea |
Drought increased Trans-2-hexenal, phytol, and δ-tocopherol |
[80] |
||||
|
Melissa officinalis |
Ozone treatment increased Rosmarinic acid, Anthocynine, Tannine, and Antioxidant activity |
[81] |
||||
|
Lycopersicon esculentum |
Soil nutrients, Nitrogen and phosphate increased Quercetin |
[82] |
||||
|
Nanoparticle (Nano-elicitor) |
Silver nanoparticles (AgNPs), Zinc oxide nanoparticles (ZnO NPs) |
Modulates redox signaling and enzyme activation related to secondary metabolism |
Phenolics, terpenes, alkaloids |
Stevia rebaudiana Bert |
Ag increased Rebaudioside-A content |
[83] |
|
Lilium ledebourii |
ZnO increased Total phenolic, Flavonoid and Anthocyanin |
[84] |
||||
|
Dracocephalum moldavica |
TiO2 increased Luteolin 7- O-glucoside, Rosmarinic acid, P-cumaric acid, Ellagitannin, Gentisic, Chlorogenic acid, and Caffeic acid |
[85] |
||||
|
Lepidum sativum |
Fe3O4 increased Essential oil, Phenolic and Flavonoid contents. |
[86] |
||||
|
Stevia rebaudiana Bertoni |
Cu-Au increased Total phenolic, Flavonoid contents |
[87] |
||||
|
Withania somnifera |
Ag +Zn increased Withanolide |
[88] |
||||
|
Plant hormone elicitors |
Salicylic acid (SA), Abscisic acid (ABA) |
Activates stress-responsive transcription and secondary metabolism genes |
Phenolics, glycosides |
Hemerocallis citrina |
Salicylic acid-induced higher accumulation of colchicine, ascorbic acid, soluble protein and soluble sugar |
[89] |
Table 1: Overview of chemical, biotic, abiotic, nano, and hormonal elicitors reported to enhance secondary metabolite biosynthesis in horticultural crops
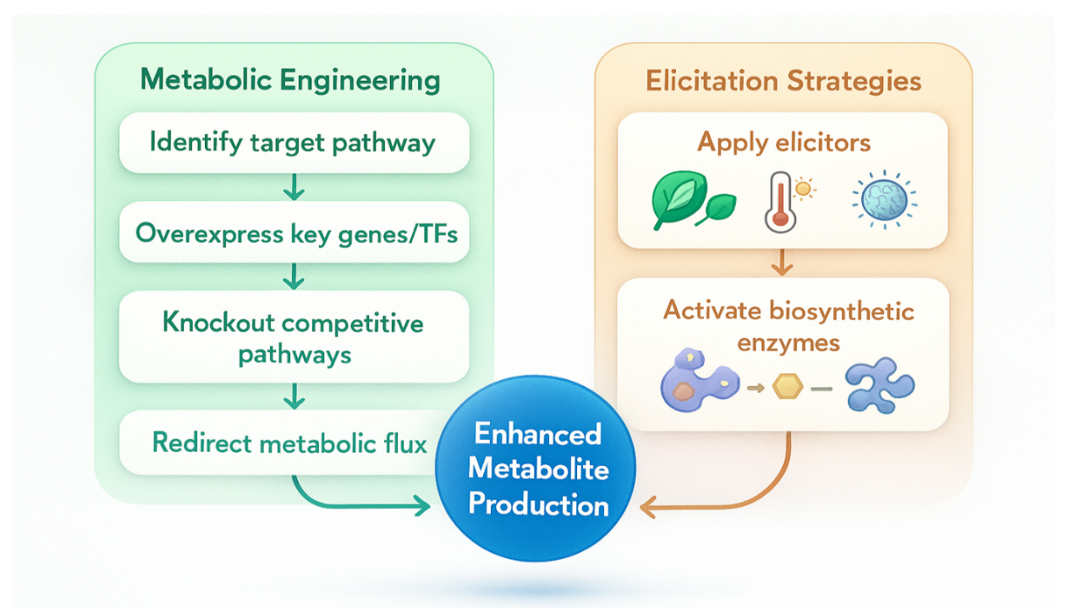
Figure 1: Integrated framework of metabolic engineering and elicitation strategies for enhanced secondary metabolite production in horticultural crops
The model demonstrates that metabolic engineering and elicitation function synergistically to optimize the biosynthesis of secondary metabolites. The metabolic engineering module includes the identification of key biosynthetic pathways, overexpression of structural or regulatory genes such as transcription factors, elimination of competing branches, and redirection of metabolic flux toward desired metabolites. The elicitation module involves the application of biotic, abiotic, nano-, or hormonal elicitors, which activate biosynthetic enzymes via signal transduction and defense pathways. Collectively, these strategies generate a coordinated molecular and physiological response that improves metabolite yield, stability, and productivity in horticultural systems.
Integration of Metabolic Engineering and Elicitation Strategies
A synergistic approach that combines genetic engineering with elicitation has become a powerful and sustainable method for enhancing secondary metabolite production in horticultural crops. Genetic engineering stably increases biosynthetic capacity by overexpressing key enzymes or transcription factors. In contrast, elicitation introduces dynamic signaling stimuli, such as jasmonates, salicylic acid, or chitosan, which activate these pathways in a temporally responsive manner. Recent studies indicate that the combination of these strategies produces synergistic effects on metabolite accumulation [90]. For instance, in Solanum lycopersicum (tomato), overexpression of rate-limiting enzymes in the carotenoid pathway, including 1-deoxy-D-xylulose-5-phosphate synthase (DXS) and Phytoene Synthase (PSY), together with methyl jasmonate (MeJA) elicitation, significantly increased carotenoid yields [91]. The application of MeJA has also been shown to enhance the production of bioactive compounds such as phenolics and flavonoids, which are essential for antioxidant properties, thereby improving the health benefits of crops like Ocimum tenuiflorum (basil) and Solanum lycopersicum (tomato) [92]. Furthermore, non-transgenic approaches to increase specific antioxidant levels, such as carnosic acid in rosemary, underscore the importance of understanding biosynthetic pathways to optimize metabolite profiles without altering genetic integrity [93]. In aromatic crops such as Ocimum basilicum, MeJA treatment combined with transcription factor overexpression (e.g., WRKY) has resulted in two- to tenfold increases in total phenolic and terpenoid content [92,94]. This integrated strategy provides both a robust genetic foundation and an inducible regulatory mechanism, thereby improving the yield, stability, and bioactivity of valuable phytochemicals in horticultural systems.
Establishing an operational framework for integrating metabolic engineering and elicitation in horticultural crops is essential to facilitate rational, data-driven decision-making. A four-phase conceptual model is proposed, linking target definition, pathway diagnosis, strategic intervention, and translational validation. In Phase I, target metabolites of agronomic or commercial interest are identified, and pathway fluxes are evaluated using transcriptomic and metabolomic data. Phase II involves diagnosing bottlenecks through enzyme-activity profiling or precursor-flux analysis to identify rate-limiting steps. In Phase III, strategy selection is based on the characterization of pathway genes or regulatory transcription factors (e.g., MYB, bHLH, or AP2/ERF families), with metabolic engineering prioritized when these elements are well defined, and elicitation favored for activating latent defense-related pathways or enhancing secondary metabolite pools in cultured tissues. Phase IV includes optimization, scale-up, and regulatory assessment to ensure reproducibility from laboratory to controlled-environment or field systems. This framework enables integration by aligning predictive metabolic rewiring with responsive elicitation, thereby maximizing biosynthetic efficiency while minimizing unintended trade-offs. Comparable holistic workflows have been recommended for medicinal and aromatic crops and are increasingly supported by omics-assisted modeling approaches [95-98]. Formalizing these stages allows horticultural researchers to systematically select strategies that are scientifically justified as well as economically and environmentally scalable.
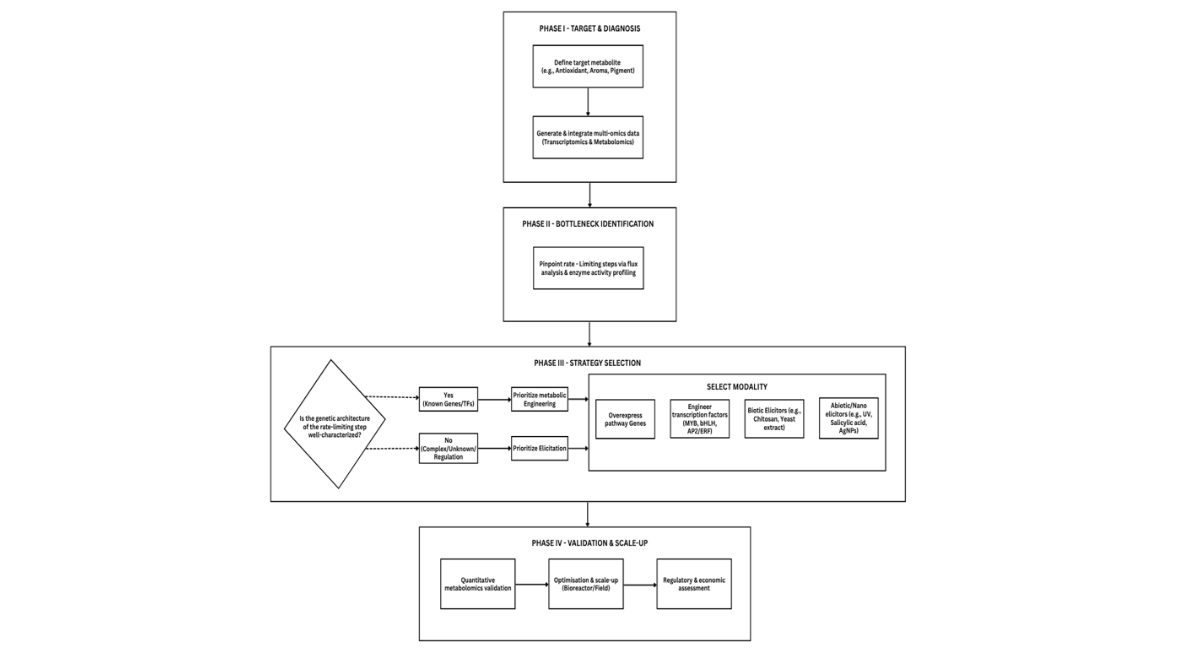
Figure 2: Operational framework for integrating metabolic engineering and elicitation strategies in horticultural crops
This systematic, four-phase model provides a structured approach from target identification to scalable validation. In Phase I, target metabolites of commercial or agronomic significance are defined, and pathway fluxes are assessed using integrated transcriptomic and metabolomic analyses. Phase II involves diagnosing bottlenecks through enzyme activity profiling and precursor flux analysis to identify rate-limiting steps. In Phase III, strategy selection is informed by biological knowledge: metabolic engineering is prioritized when pathway genes or regulatory transcription factors (TFs; such as MYB, bHLH, AP2/ERF) are well-characterized, while elicitation is preferred for activating latent defense-related pathways or when genetic information is limited. Both strategies may be combined as appropriate. Phase IV focuses on optimizing and validating promising interventions through quantitative metabolomics, followed by scale-up and regulatory assessment to ensure both economic and environmental scalability.
Quantitative Case Studies in Horticultural Crop Biotechnology
The integration of metabolic engineering with exogenous elicitation is increasingly employed to address regulatory and precursor-supply limitations in secondary-metabolite production within horticultural crops. Genetic engineering enhances pathway capacity, while elicitors such as methyl jasmonate (MeJA) activate stress-responsive signaling that redirects metabolic flux. Quantitative omics and metabolite analyses indicate that this combined approach is highly effective. For example, fruit-specific expression of an MYB flavonol regulator in tomato significantly increases flavonol content: AtMYB12/SlMYB12 expression in tomato fruits resulted in approximately 10- to 12-fold increases in rutin and related flavonols in transgenic lines [99]. In isoprenoid pathways, augmenting precursor supply through 1-deoxy-D-xylulose-5-phosphate synthase (DXS) expression has consistently elevated carotenoid levels. Constitutive DXS overexpression in heterologous plant systems produced 1.7- to 2.1-fold increases in total carotenoids and specific carotenes, highlighting the importance of precursor engineering as a component of integrated strategies [100,101]. In medicinal plant systems where jasmonate signaling is pivotal, transcriptional engineering of jasmonic acid (JA)-responsive regulators yields substantial improvements. Overexpression of the bHLH factor CrMYC1 in Catharanthus roseus hairy-root lines increased ajmalicine by approximately 13- to 14-fold and catharanthine by 3- to 4.4-fold, as quantified by HPLC. The JA-responsive CrMYC/ORCA regulatory network mediates MeJA-induced TIA biosynthesis, demonstrating that the combination of transcription factor-based engineering with jasmonate elicitation or other elicitors can produce synergistic, super-additive increases in target alkaloids [102,103].
Table 2 presents key case studies that quantitatively support this synergistic strategy across diverse horticultural species and metabolite classes. Collectively, these findings indicate that integrating transcriptional or enzymatic modulation with targeted elicitation establishes a positive feedback loop, thereby unlocking the full biosynthetic potential of plant systems and expediting the translation of research into commercially viable horticultural biotechnology.
|
Horticultural crop / system |
Target metabolite(s) |
Intervention strategy |
Key quantitative outcomes |
Reference |
|
Solanum lycopersicum (tomato; Del/Ros1 transgenic lines) |
Anthocyanins (fruit peel and flesh) |
Genetic modification: fruit-specific overexpression of Delila (Del) and Rosea1 (Ros1) transcription factors using the E8 promoter |
Transgenic lines produced deeply pigmented fruits with total anthocyanin concentrations comparable to blackberry/blueberry; hydrophilic antioxidant capacity increased approximately threefold relative to wild-type controls (HPLC-based quantification). |
[104] |
|
Catharanthus roseus (hairy-root cultures) |
Terpenoid indole alkaloids (ajmalicine, catharanthine) |
Genetic modification: overexpression of CrMYC1 transcription factor with or without MeJA elicitation |
CrMYC1-overexpressing lines accumulated ajmalicine ~13–14-fold and catharanthine ~3–4.4-fold higher than controls (HPLC quantification). |
[105] |
|
Vitis vinifera cv. Monastrell (cell-suspension cultures) |
Stilbenes (trans-resveratrol and derivatives) |
Elicitation: methyl jasmonate (MeJA) combined with β-cyclodextrin (CD) |
MeJA + CD treatment increased extracellular trans-resveratrol and total stilbenes from ~25 mg L⁻¹ (control) to ~100–130 mg L⁻¹, depending on induction conditions. |
[106] |
|
Capsicum spp. (cell cultures and fruit tissues) |
Capsaicinoids (capsaicin, dihydrocapsaicin) |
Elicitation: MeJA (varied doses) with additional influences from sucrose/media composition, cultivar genotype, and TF regulation (e.g., CaMYB108) |
Multiple studies report significant MeJA-induced increases in capsaicinoid accumulation, with multi-fold enhancements in both in vitro cultures and fruits, alongside increased PAL activity and upregulation of capsaicinoid biosynthetic genes. Specific fold-change values vary by species, cultivar, and elicitation regime. |
|
|
Ocimum basilicum (basil; hairy-root or callus cultures) |
Rosmarinic acid (RA) and total phenolics |
Elicitation with abiotic inducers (e.g., chitosan) in hairy-root or callus platforms |
Elicited hairy-root/callus lines commonly show 2–4-fold increases in RA relative to non-elicited controls; some transformed root lines reach ~10 mg g⁻¹ FW (≈1% FW) RA content depending on culture conditions. |
[109] |
|
Brassica oleracea var. italica (broccoli; seedlings, hairy roots, or pre-harvest treatments) |
Glucoraphanin (precursor of sulforaphane) |
Elicitation: MeJA foliar application or MeJA induction in hairy-root cultures |
MeJA treatment consistently enhances glucoraphanin levels, with reported multi-fold increases relative to untreated controls; transcriptomic analyses show induction of key biosynthetic genes including CYP79F1/2. Magnitude varies with dose, genotype, and sampling time. |
[110] |
Table 2: Quantitative outcomes of integrated metabolic engineering and elicitation strategies in horticultural crops
Integration of Omics Data and Systems Biology Perspectives
The integration of omics data has significantly advanced the understanding of complex biological systems and facilitated progress in personalized medicine. High-throughput technologies generate extensive datasets, enabling the identification of biomarkers essential for tailoring therapies to individual patients, which improves treatment efficacy and reduces adverse effects [111]. The interaction among genomics, transcriptomics, proteomics, and metabolomics provides insights into the dynamic processes underlying diseases and creates opportunities for novel therapeutic interventions [112]. Despite these advances, developing robust bioinformatics tools to manage and interpret multifaceted omics data remains a challenge, highlighting the ongoing need for innovation in systems biology methodologies to fully realize the potential of integrative omics [113].
Multi-omics approaches, including genomics, transcriptomics, proteomics, and metabolomics, are increasingly recognized as essential for elucidating the regulatory networks governing secondary metabolism in horticultural crops [114]. Genomics establishes the biosynthetic potential, while transcriptomics identifies gene clusters and co-expression modules activated by elicitor exposure.
Metabolomics quantifies the biochemical outputs of these networks [115], and proteomics provides information on post-translational modifications and dynamic enzyme complexes [116]. Network and systems biology methods integrate these data layers to identify bottleneck enzymes, cross-pathway regulators, and gene–metabolite relationships [32]. Advanced metabolic models simulate flux redistribution under combined metabolic engineering and elicitation scenarios, supporting the rational design of processes to maximize metabolite yield [117,118]. Collectively, these integrative methods offer a predictive framework for precision metabolic engineering, improving the capacity to translate elicitor signals and genetic modifications into consistent increases in phytochemical production.
Addressing Translational Challenges: Advancing from Laboratory Research to Field and Commercial Applications
Although elicitation and metabolic engineering have demonstrated success in controlled environments, translating laboratory-scale advances to commercial horticultural systems remains challenging. A primary obstacle is the scale-dependent variability in metabolite yield. Elicitor concentrations, nutrient dynamics, and microenvironmental cues optimized in vitro often do not translate effectively to field or bioreactor conditions [119]. Bioprocess scale-up introduces additional challenges, including oxygen transfer limitations, shear stress, and metabolite feedback inhibition, all of which can compromise process stability and reproducibility [120]. To address these issues, bioreactor-based optimization strategies, such as airlift and stirred-tank systems with integrated online monitoring, have been developed to enable real-time elicitor control and metabolite tracking [121]. In addition, multi-omics-driven process modeling supports the prediction of metabolic flux redistribution under dynamic culture conditions, facilitating rational parameter optimization [122]. From a regulatory perspective, the deployment of engineered horticultural crops or microbial chassis is constrained by concerns regarding biosafety, environmental release, and consumer acceptance [123,124]. Therefore, harmonized risk assessment frameworks and transparent data-sharing protocols are essential for the safe translation of laboratory-engineered systems into practical applications. Ultimately, overcoming these translational barriers through scalable bioprocessing platforms, regulatory harmonization, and techno-economic modeling will determine the sustainable commercialization of metabolite-enriched horticultural products.
Prospective Developments
Future advancements in enhancing secondary metabolite production in horticultural crops depend on transitioning from descriptive elicitation experiments to data-driven, predictive, and scalable frameworks. Integrating multi-omics technologies—genomics, transcriptomics, proteomics, and metabolomics—with systems biology modeling can unravel key regulatory nodes controlling metabolite fluxes and enable rational selection of engineering and elicitation strategies [125]. Artificial intelligence (AI) and machine learning approaches are emerging as powerful tools to analyze large omics datasets and predict elicitor–response dynamics, thereby facilitating real-time optimization of elicitor combinations and concentrations in bioreactor systems [126,127]. In parallel, CRISPR/Cas-based multiplex genome editing offers the potential to precisely modify multiple pathway genes simultaneously, overcoming feedback inhibition and enabling coordinated control of biosynthetic flux [128]. These genetic interventions can be coupled with inducible synthetic promoters and transcription factors for fine-tuned, on-demand metabolite synthesis.
Translational progress will also require scalable production systems such as temporary immersion bioreactors and controlled elicitor delivery platforms, which provide reproducibility and environmental stability during metabolite accumulation [129]. Furthermore, adopting nanocarrier-mediated elicitor formulations can enhance bioavailability and uptake efficiency, improving elicitation responsiveness under field or in vitro conditions. However, regulatory and societal barriers surrounding genetically modified horticultural products remain critical challenges, necessitating a shift toward cisgenic or transgene-free genome-edited plants with clear biosafety compliance[123].
Finally, sustainability-driven innovations such as plant cell-free systems and microbial chassis (e.g., engineered Saccharomyces cerevisiae or Escherichia coli) expressing plant biosynthetic pathways offer promising alternatives for high-yield metabolite production under controlled, environment-independent conditions [130]. Collectively, integrating CRISPR-based metabolic engineering, multi-omics-guided network modeling, and AI-assisted elicitation strategies—alongside scalable, regulatory-compliant production systems will redefine the future of horticultural metabolite biotechnology, enabling predictable, sustainable, and commercially viable synthesis of high-value phytochemicals.
CONCLUSION
The integration of metabolic engineering and elicitation strategies marks a significant advancement in horticultural biotechnology, facilitating a shift from empirical enhancement of secondary metabolites to predictable, system-driven optimization. This review demonstrates that, although each strategy independently offers considerable potential, their combination fundamentally redefines the regulation of plant metabolic fluxes. Analysis of current literature indicates that elicitation induces temporal activation of defense-related biosynthetic pathways via stress signaling, while metabolic engineering provides sustained, structural modification of gene networks supporting metabolite accumulation. Nevertheless, many studies remain primarily descriptive, reporting increases in metabolite levels without establishing correlations to pathway kinetics, transcriptional regulation, or physiological trade-offs.
A primary limitation within the field is the lack of standardization and cross-comparative quantification. Although recent progress in omics technologies and CRISPR-mediated genome editing has enabled precise pathway modification, challenges persist regarding scalability, stability, and reproducibility in field environments. Additionally, the predominant use of short-term in vitro models restricts understanding of elicitor–gene interactions under variable environmental conditions, soil microbiome dynamics, and photoperiodic changes. Consequently, a significant gap remains in translating laboratory-scale biochemical achievements into agronomic performance and economic viability.
Overcoming these challenges requires the development of an operational, data-driven framework that incorporates pathway bottleneck analysis, regulatory target identification, elicitor selection, and quantitative metabolomic validation. This approach would transform elicitation from an empirical process into a rationally designed intervention, customized for specific crop types, target metabolites, and production scales. Concurrently, systems biology models can simulate metabolic flux redistribution under combined interventions, supporting prediction-based experimental design in place of traditional trial-and-error methods.
Looking ahead, the integration of multi-omics approaches, artificial intelligence-assisted modeling, and modular genetic control is expected to shape the next phase of horticultural metabolic research. The primary challenge will extend beyond increasing secondary metabolite levels to achieving production systems that are stable, scalable, and responsive to environmental conditions. Lasting innovation will depend on the creation of integrative frameworks that combine molecular precision, physiological adaptability, and translational feasibility, thereby bridging the persistent gap between laboratory potential and field application.
ABBREVIATIONS
TF, Transcription Factor; AgNPs, Silver Nanoparticles.
REFERENCES
- Tang H, Wang Q, Xie H, Li W. The function of secondary metabolites in resisting stresses in horticultural plants. Fruit Res. 2024;4(1):0. [Google Scholar]
- Xu L, Wang X. A comprehensive review of phenolic compounds in horticultural plants. Int J Mol Sci. 2025;26(12):5767. [Crossref] [Google Scholar] [PubMed]
- Khanam S, Mishra P, Faruqui T, Alam P, Albalawi T, et al. Plant-based secondary metabolites as natural remedies: a comprehensive review on terpenes and their therapeutic applications. Front Pharmacol. 2025;16:1587215. [Crossref] [Google Scholar] [PubMed]
- Goura K, Legrifi I, Kallali NS, Taoussi M, Kenfaoui J, et al. Beyond Survival: The role of secondary metabolites in plant defense mechanisms. J Crop Health. 2025;77(4). [Crossref] [Google Scholar]
- Ozyigit II, Dogan I, Hocaoglu-Ozyigit A, Yalcin B, Erdogan A, et al. Production of secondary metabolites using tissue culture-based biotechnological applications. Front Plant Sci. 2023;14:1132555. [Crossref] [Google Scholar] [PubMed]
- Pang Z, Chen J, Wang T, Gao C, Li Z, Guo L, et al. Linking plant secondary metabolites and plant microbiomes: A review. Front Plant Sci. 2021;12:621276. [Crossref] [Google Scholar] [PubMed]
- Singh KS, Van Der Hooft JJJ, Van Wees SCM, Medema MH. Integrative omics approaches for biosynthetic pathway discovery in plants. Nat Prod Rep. 2022;39(9):1876–96. [Crossref] [Google Scholar] [PubMed]
- Fazili MA, Bashir I, Ahmad M, Yaqoob U, Geelani SN. In vitro strategies for the enhancement of secondary metabolite production in plants: a review. Bull Natl Res Cent. 2022;46(1):41. [Crossref] [Google Scholar] [PubMed]
- Karakas E, Bulut M, Fernie AR. The use of web resources for metabolomics in horticultural crops. Hortic Adv. 2025;3(1):12. [Crossref] [Google Scholar]
- Verpoorte R, Contin A, Memelink J. Biotechnology for the production of plant secondary metabolites. Phytochemistry reviews. 2002; 1(1):13-25. [Crossref] [Google Scholar]
- Singh S, Chhatwal H, Pandey A. Deciphering the complexity of terpenoid biosynthesis and its multi-level regulatory mechanism in plants. J Plant Growth Regul. 2024;43(10):3320–36. [Crossref] [Google Scholar]
- Pratyusha DS, Sarada DVL. MYB transcription factors—master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 2022;41(12):2245–60. [Crossref] [Google Scholar] [PubMed]
- Walters KJ, Lopez RG, Behe BK. Leveraging Controlled-Environment agriculture to increase key basil terpenoid and phenylpropanoid concentrations: the effects of radiation intensity and CO2 concentration on consumer preference. Front Plant Sci. 2021;11:598519. [Crossref] [Google Scholar] [PubMed]
- Fuchs LK, Holland AH, Ludlow RA, Coates RJ, Armstrong H, et al. Genetic manipulation of biosynthetic pathways in Mint. Front Plant Sci. 2022;13:928178. [Crossref] [Google Scholar] [PubMed]
- Zhang Y, Li Y, Li W, Hu Z, Yu X, et al. Metabolic and molecular analysis of nonuniform anthocyanin pigmentation in tomato fruit under high light. Hortic Res. 2019;6(1):26. [Crossref] [Google Scholar] [PubMed]
- Manners GD. Citrus limonoids: analysis, bioactivity, and biomedical prospects. J Agric Food Chem. 2007;55(21):8285–94. [Crossref] [Google Scholar] [PubMed]
- Reyes-Escogido MD, Gonzalez-Mondragon EG, Vazquez-Tzompantzi E. Chemical and pharmacological aspects of capsaicin. Molecules. 2011;16(2):1253–70. [Google Scholar] [PubMed]
- Rajkumari S, Sanatombi K. In Vitro Production of Some Important Secondary Metabolites from Zingiber Species. In: Jha S, editor. Medicinal Aromatic Plant Sci. Biotech. Singapore: Springer; 2018. p. 213–33. [Crossref] [Google Scholar]
- Biswas D, Chakraborty A, Mukherjee S, Ghosh B. Hairy root culture: a potent method for improved secondary metabolite production of Solanaceous plants. Front Plant Sci. 2023;14:1197555. [Crossref] [Google Scholar] [PubMed]
- Rinaldi MA, Ferraz CA, Scrutton NS. Alternative metabolic pathways and strategies to high-titre terpenoid production in Escherichia coli. Nat Prod Rep. 2021;39(1):90–118. [Crossref] [Google Scholar] [PubMed]
- Lim W, Li J. Co-expression of onion chalcone isomerase in Del/Ros1-expressing tomato enhances anthocyanin and flavonol production. Plant Cell Tissue Organ Cult. 2016;128(1):113–124. [Crossref] [Google Scholar]
- Fuchs LK, Holland AH, Ludlow RA, Coates RJ, Armstrong H, et al. Genetic manipulation of biosynthetic pathways in Mint. Front Plant Sci. 2022;13:928178. [Crossref] [Google Scholar] [PubMed]
- Xu C, Fan X, Shen G, Guo B. Genome-wide identification of the phenylalanine ammonia-lyase gene from Epimedium Pubescens Maxim. (Berberidaceae): novel insight into the evolution of the PAL gene family. BMC Plant Biol. 2024;24(1):482. [Crossref] [Google Scholar] [PubMed]
- Yan T, Shu X, Ning C, Li Y, Wang Z, et al. Functions and Regulatory Mechanisms of bHLH Transcription Factors during the Responses to Biotic and Abiotic Stresses in Woody Plants. Plants. 2024;13(16):2315. [Crossref] [Google Scholar] [PubMed]
- Li H, Chen N, Zhang H, Xu D. Multidimensional regulation of transcription factors: decoding the comprehensive signals of plant secondary metabolism. Front Plant Sci. 2025;16:1522278. [Crossref] [Google Scholar] [PubMed]
- Pandey A, Misra P, Choudhary D, Yadav R, Goel R, et al. AtMYB12 expression in tomato leads to large scale differential modulation in transcriptome and flavonoid content in leaf and fruit tissues. Sci Rep. 2015;5:12412. [Crossref] [Google Scholar] [PubMed]
- Cao Y, Li K, Li Y, Zhao X, Wang L. MYB transcription factors as regulators of secondary metabolism in plants. Biology. 2020;9(3):61. [Crossref] [Google Scholar] [PubMed]
- Zhou W, Shi M, Deng C, Lu S, Huang F, et al. The methyl jasmonate-responsive transcription factor SmMYB1 promotes phenolic acid biosynthesis in Salvia miltiorrhiza. Hortic Res. 2021;8(1):25. [Crossref] [Google Scholar] [PubMed]
- Tong Y, Xue J, Li Q, Zhang L. A generalist regulator: MYB transcription factors regulate active ingredient biosynthesis in medicinal plants. J Exp Bot. 2024;75(16):4729–44. [Crossref] [Google Scholar] [PubMed]
- Mariyam S, Kumar V, Roychoudhury A, Ghodake GS, Muneer S, et al. Functional diversification and mechanistic insights of MYB transcription factors in mediating plant growth and development, secondary metabolism, and stress responses. J Plant Growth Regul. 2025; [Epub ahead of print]. [Crossref] [Google Scholar]
- Rabeh K, Hnini M, Oubohssaine M. A comprehensive review of transcription factor-mediated regulation of secondary metabolites in plants under environmental stress. Stress Biol. 2025;5(1):15. [Crossref] [Google Scholar]
- Zhao Y, Liu G, Yang F, Liang Y, Gao Q, et al. Multilayered regulation of secondary metabolism in medicinal plants. Mol Hortic. 2023;3(1):11. [Crossref] [Google Scholar] [PubMed]
- Daniel MA, Sebastin R, Yu JK, Soosaimanickam MP, Chung JW. Enhancing Horticultural Crops through Genome Editing: Applications, Benefits, and Considerations. Horticulturae. 2023;9(8):884. [Crossref] [Google Scholar]
- Rukavtsova EB, Zakharchenko NS, Lebedev VG, Shestibratov KA. CRISPR-CAS Genome Editing for Horticultural Crops Improvement: Advantages and Prospects. Horticulturae. 2022;9(1):38. [Crossref] [Google Scholar]
- Sunnadeniya R, Bean A, Brown M, Akhavan N, Hatlestad G, et al. Tyrosine Hydroxylation in Betalain Pigment Biosynthesis Is Performed by Cytochrome P450 Enzymes in Beets (Beta vulgaris). PLoS One. 2016;11(2):e0149417. [Crossref] [Google Scholar] [PubMed]
- Ogawa K, Murota K, Shimura H, Furuya M, Togawa Y, et al. Evidence of capsaicin synthase activity of the Pun1-encoded protein and its role as a determinant of capsaicinoid accumulation in pepper. BMC Plant Biol. 2015;15:93. [Crossref] [Google Scholar] [PubMed]
- Qin K, Liu F, Zhang C, Deng R, Fernie AR, Zhang Y. Systems and synthetic biology for plant natural product pathway elucidation. Cell Rep. 2025;44(6):115715. [Crossref] [Google Scholar] [PubMed]
- Li Q, Sapkota M, Van Der Knaap E. Perspectives of CRISPR/Cas-mediated cis-engineering in horticulture: unlocking the neglected potential for crop improvement. Hortic Res. 2020;7:36. [Crossref] [Google Scholar] [PubMed]
- Bai W, Li C, Li W, Wang H, Han X, et al. Machine learning assists prediction of genes responsible for plant specialized metabolite biosynthesis by integrating multi-omics data. BMC Genomics. 2024;25(1):384. [Crossref] [Google Scholar] [PubMed]
- Gao Y, Zhao C. Development and applications of metabolic models in plant multi-omics research. Front Plant Sci. 2024;15:1361183. [Crossref] [Google Scholar] [PubMed]
- Rao X, Liu W. A Guide to Metabolic network modeling for Plant Biology. Plants. 2025;14(3):484. [Crossref] [Google Scholar] [PubMed]
- Singh KS, Duran HS, Del Pup E, Delgado OZ, Van Wees SCM, et al. MEANtools integrates multi-omics data to identify metabolites and predict biosynthetic pathways. PLoS Biol. 2025;23(7):e3003307. [Crossref] [Google Scholar] [PubMed]
- Pomahačová B, Dušek J, Dušková J, Yazaki K, Roytrakul S, et, al. Improved accumulation of ajmalicine and tetrahydroalstonine in Catharanthus cells expressing an ABC transporter. J Plant Physiol. 2009;166(13):1405–12. [Crossref] [Google Scholar] [PubMed]
- Shoji T. ATP-Binding cassette and multidrug and toxic compound extrusion transporters in plants. Int Rev Cell Mol Biol. 2014;309:303–46. [Crossref] [Google Scholar] [PubMed]
- Zhao J, Dixon RA. MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-Glucoside for proanthocyanidin biosynthesis in Medicago Truncatula and Arabidopsis. Plant Cell. 2009;21(8):2323–40. [Crossref] [Google Scholar] [PubMed]
- Xia X, Fan X, Wei J, Feng H, Qu H, et al. Rice nitrate transporter OsNPF2.4 functions in low-affinity acquisition and long-distance transport. J Exp Bot. 2014;66(1):317–31. [Crossref] [Google Scholar] [PubMed]
- Cao H, Nuruzzaman M, Xiu H, Huang J, Wu K, et al. Transcriptome Analysis of Methyl Jasmonate-Elicited Panax ginseng Adventitious Roots to Discover Putative Ginsenoside Biosynthesis and Transport Genes. Int J Mol Sci. 2015;16(2):3035–57. [Crossref] [Google Scholar] [PubMed]
- Vezzulli S, Civardi S, Ferrari F, Bavaresco L. Methyl jasmonate treatment as a trigger of resveratrol synthesis in cultivated grapevine. Am J Enol Vitic. 2007;58(4):530–3. [Crossref] [Google Scholar]
- Li Z, Wang X, Chen F, Kim HJ. Chemical Changes and Overexpressed Genes in Sweet Basil (Ocimum basilicum L.) upon Methyl Jasmonate Treatment. J Agric Food Chem. 2007;55(3):706–13. [Crossref] [Google Scholar] [PubMed]
- Chowdhury MR, Mehmet M, Mukherjee J, Debnath AJ, Ražná K. Chitosan as an Elicitor in Plant Tissue Cultures: Methodological challenges. Molecules. 2025;30(17):3476. [Crossref] [Google Scholar] [PubMed]
- Bhaskar R, Xavier LSE, Udayakumaran G, Kumar DS, Venkatesh R, et, al. Biotic elicitors: a boon for the in-vitro production of plant secondary metabolites. Plant Cell Tissue Organ Cult. 2021;149(1–2):7–24. [Crossref] [Google Scholar]
- Jain D, Bisht S, Parvez A, Singh K, Bhaskar P, et, al. Effective biotic elicitors for augmentation of secondary metabolite production in medicinal plants. Agriculture. 2024;14(6):796. [Crossref] [Google Scholar]
- Martínez-Zamora L, Castillejo N, Artés-Hernández F. UV-B radiation as abiotic elicitor to enhance phytochemicals and development of red cabbage sprouts. Horticulturae. 2021;7(12):567. [Crossref] [Google Scholar]
- Emus-Medina A, Contreras-Angulo LA, Ambriz-Perez DL, Vazquez-Olivo G, Heredia JB. UV Light Stress Induces Phenolic Compounds in Plants. Plant Stress: Challenges and Management in the New Decade. Cham: Springer; 2023. p. 415–40. [Crossref] [Google Scholar]
- Kim CK, Eom SH. Light controls in the regulation of carotenoid biosynthesis in leafy vegetables: a review. Horticulturae. 2025;11(2):152. [Crossref] [Google Scholar]
- De Jesus Rivero-Montejo S, Vargas-Hernandez M, Torres-Pacheco I. Nanoparticles as novel elicitors to improve bioactive compounds in plants. Agriculture. 2021;11(2):134. [Crossref] [Google Scholar]
- Lala S. Nanoparticles as elicitors and harvesters of economically important secondary metabolites in higher plants: A review. IET Nanobiotechnol. 2021;15(1):28–57. [Crossref] [Google Scholar] [PubMed]
- Shahin H. Enhanced production of secondary metabolites by methyl jasmonate and silver nanoparticles elicitation in tissue culture of Catharanthus roseus (Apocynaceae). Al-Azhar J Pharm Sci. 2018;57(1):62–9. [Crossref] [Google Scholar]
- Humbal A, Pathak B. Harnessing nanoparticle-mediated elicitation in plant tissue culture: a promising approach for secondary metabolite production. Plant Cell Tissue Organ Cult. 2023;155(2):385–402. [Crossref] [Google Scholar]
- Aziz MA, Shamim A, Masmoudi K. Pattern recognition receptors in plant immunity. Adv Exp Med Biol. 2025;1411:425–51. [Crossref] [Google Scholar] [PubMed]
- Kong F, Ramonell KM. Receptor-like kinases and their role in plant innate immunity. In: Goyal A, editor. Effector-Triggered Immunity. Academic Press; 2022. p.39–62. [Crossref] [Google Scholar]
- Ho TT, Murthy HN, Park SY. Methyl jasmonate induced oxidative stress and accumulation of secondary metabolites in plant cell and organ cultures. Int J Mol Sci. 2020;21(3):716. [Crossref] [Google Scholar] [PubMed]
- Jeyasri R, Muthuramalingam P, Karthick K, Shin H, Choi SH, et, al. Methyl jasmonate and salicylic acid as powerful elicitors for enhancing the production of secondary metabolites in medicinal plants: an updated review. Plant Cell Tissue Organ Cult. 2023;153(3):447–58. [Crossref] [Google Scholar] [PubMed]
- Haghpanah M, Namdari A, Kaleji MK, Nikbakht-Dehkordi A, Arzani A, et, al. Interplay between ROS and hormones in plant defense against pathogens. Plants. 2025;14(9):1297. [Crossref] [Google Scholar] [PubMed]
- Wasternack C, Song S. Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transciption. J Exp Bot. 2016;68(6):1303–21. [Crossref] [Google Scholar] [PubMed]
- Lukan T, Coll A. Intertwined roles of reactive oxygen species and salicylic acid signaling are crucial for the plant response to biotic stress. Int J Mol Sci. 2022;23(10):5568. [Crossref] [Google Scholar] [PubMed]
- Kantayos V, Kim JS, Baek SH. Alteration of resveratrol-dependent glycosyltransferase activity by elicitation in DJ-526 rice. GM Crops Food. 2021;12(1):242–50. [Crossref] [Google Scholar] [PubMed]
- Pillai SK, Siril EA. Exogenous Elicitors Enhanced Berberine Production in the Cell Suspension Cultures of Tinospora cordifolia (Willd.) Miers ex Hook F. &Thoms. Proc Natl Acad Sci India Sect B Biol Sci. 2022;92(1):209–18. [Crossref] [Google Scholar]
- Rajan M, Feba KS, Chandran V, Shahena S, Mathew L. Enhancement of rhamnetin production in Vernonia anthelmintica (L.) Willd. cell suspension cultures by eliciting with methyl jasmonate and salicylic acid. Physiol Mol Biol Plants. 2020;26(7):1531–9. [Crossref] [Google Scholar] [PubMed]
- Dasari R, Gopu C, Vankudoth S, Dharavath S, Taduri S. Enhancement of production of pharmaceutically important anti-cancerous compound; cucurbitacin E via elicitation and precursor feeding of in vitro culture of Citrullus colocynthis (L.) Schard. Vegetos. 2020;33(2):323–34. [Crossref] [Google Scholar]
- Razavizadeh R, Adabavazeh F. In vitro application of chitosan effects on essential oil content and physiological characteristics of Dracocephalum kotschyi Boiss. J Plant Process Funct. 2019;8(31):23–30. [Google Scholar]
- Dar TA, Uddin M, Khan MMA, Ali A, Mir SR, et, al. Effect of Co-60 gamma irradiated chitosan and phosphorus fertilizer on growth, yield and trigonelline content of Trigonella foenum-graecum L. J Radiat Res Appl Sci. 2015;8(3):446–58. [Crossref] [Google Scholar]
- Zaker A, Sykora C, Gössnitzer F, Abrishamchi P, Asili J, et al. Effects of some elicitors on tanshinone production in adventitious root cultures of Perovskia abrotanoides Karel. Ind Crops Prod. 2015;67:97–102. [Crossref] [Google Scholar]
- Naik PM, Al-Khayri JM. Abiotic and Biotic Elicitors–Role in Secondary Metabolites Production through In Vitro Culture of Medicinal Plants. London: IntechOpen; 2016. [Crossref] [Google Scholar]
- Simic SG, Tusevski O, Maury S, Delaunay A, Joseph C, et, al. Effects of polysaccharide elicitors on secondary metabolite production and antioxidant response in Hypericum perforatum L. Shoot cultures. ScientificWorldJournal. 2014;2014:609649. [Crossref] [Google Scholar] [PubMed]
- Awad V, Kuvalekar A, Harsulkar A. Microbial elicitation in root cultures of Taverniera cuneifolia (Roth) Arn. for elevated glycyrrhizic acid production. Ind Crops Prod. 2014;54:13–6. [Crossref] [Google Scholar]
- Müller V, Albert A, Winkler JB, Lankes C, Noga G, et, al. Ecologically relevant UV-B dose combined with high PAR intensity distinctly affect plant growth and accumulation of secondary metabolites in leaves of Centella asiatica L. Urban. J Photochem Photobiol B. 2013;127:161–9. [Crossref] [Google Scholar] [PubMed]
- Naghiloo S, Movafeghi A, Delazar A, Nazemiyeh H, Asnaashari S, et, al. Ontogenetic Variation of Total Phenolics and Antioxidant Activity in Roots, Leaves and Flowers of Astragalus compactus Lam. (Fabaceae). Bioimpacts. 2012;2(2):105-9. [Google Scholar] [PubMed]
- Haghighi NZ. Enhancement of compatible solute and secondary metabolites production in Plantago ovata Forsk. by salinity stress. J Med Plants Res. 2012;6(18):3525-30. [Crossref] [Google Scholar]
- Podda A, Pollastri S, Bartolini P, Pisuttu C, Pellegrini E, et al. Drought stress modulates secondary metabolites in Brassica oleracea L. convar. acephala (DC) Alef, var. sabellica L. J Sci Food Agric. 2019;99(12):5533–40. [Crossref] [Google Scholar] [PubMed]
- Tonelli M, Pellegrini E, D’Angiolillo F, Petersen M, Nali C, et al. Ozone-elicited secondary metabolites in shoot cultures of Melissa officinalis L. Plant Cell Tissue Organ Cult. 2014;120(2):617–29. [Crossref] [Google Scholar]
- Stewart AJ, Chapman W, Jenkins GI, Graham I, Martin T, et, al. The effect of nitrogen and phosphorus deficiency on flavonol accumulation in plant tissues. Plant Cell Environ. 2001;24(11):1189–97. [Crossref] [Google Scholar]
- Laha S, Subrahmanyeswari T, Verma SK, Kamble SN, Singh S, et al. Biogenic synthesis, characterization and application of silver nanoparticles as biostimulator for growth and rebaudioside-A production in genetically stable stevia (Stevia rebaudiana Bert.) under in vitro conditions. Ind Crops Prod. 2023;197:116520. [Crossref] [Google Scholar]
- Chamani E, Ghalehtaki SK, Mohebodini M, Ghanbari A. The effect of Zinc oxide nano particles and Humic acid on morphological characters and secondary metabolite production in Lilium ledebourii Bioss. Iran J Genet Plant Breed. 2015;4(2):11–9. [Google Scholar]
- Kamalizadeh M, Bihamta M, Zarei A. Drought stress and TiO2 nanoparticles affect the composition of different active compounds in the Moldavian dragonhead plant. Acta Physiol Plant. 2019;41(2):21. [Crossref] [Google Scholar]
- Ahamed TES, Ahamed ESS. Synergy Prospect Low Gamma Irradiation Doses Incorporating Elicitation with Iron Nanoparticles to Hyper Production Biomass Yield and Bioactive Secondary Metabolites for Cress, Medicinal Plant. J Plant Sci. 2018;6(5):157-65. [Google Scholar]
- Ghazal B, Saif S, Farid K, Khan A, Rehman S, et al. Stimulation of secondary metabolites by copper and gold nanoparticles in submerge adventitious root cultures of Stevia rebaudiana (Bert.). IET Nanobiotechnol. 2018;12(5):569–73. [Crossref] [Google Scholar] [PubMed]
- Singh R, Singh DP, Gupta P, Jain P, Sanchita N, et al. Nanoparticles alter the withanolide biosynthesis and carbohydrate metabolism in Withania somnifera (Dunal). Ind Crops Prod. 2018;127:94–109. [Crossref] [Google Scholar]
- Miao Y, Li H, Pan J, Zhou B, He T, et al. Salicylic acid modulates secondary metabolism and enhanced colchicine accumulation in long yellow daylily (Hemerocallis citrina). AoB Plants. 2024;16(4):plae029. [Crossref] [Google Scholar] [PubMed]
- Wang Y, Chen J, He G, Yin L, Liao Y. Unlocking the potential of flavonoid biosynthesis through integrated metabolic engineering. Front Plant Sci. 2025;16:1597007. [Crossref] [Google Scholar] [PubMed]
- Rodriguez-Concepcion M, Lim S, Ha SH. Family alliances feeding the carotenoid pathway in tomato. J Exp Bot. 2025. [Crossref] [Google Scholar]
- Chutimanukul P, Thongtip A, Panya A, Phonsatta N, Thangvichien S, et al. Enhancing the biochemical potential of holy basil through methyl jasmonate elicitation with insights into physiological responses in a plant factory. Sci Rep. 2025;15(1):20724. [Crossref] [Google Scholar] [PubMed]
- Tounekti T, Munné-Bosch S. Enhanced phenolic diterpenes antioxidant levels through non-transgenic approaches. Crit Rev Plant Sci. 2012;31(6):505–19. [Crossref] [Google Scholar]
- Li M, Shao Y, Pan B, Liu C, Tan H. Regulation of important natural products biosynthesis by WRKY transcription factors in plants. J Adv Res. 2025; [Epub ahead of print]. [Crossref] [Google Scholar]
- Zhao Y, Liu G, Yang F, Liang Y, Gao Q, et al. Multilayered regulation of secondary metabolism in medicinal plants. Mol Hortic. 2023;3(1):11. [Crossref] [Google Scholar] [PubMed]
- Novoa-del-Toro R, Witting M. Harnessing metabolites from plant cell tissue and organ culture for sustainable biotechnology. Plant Cell Tissue Organ Cult. 2025. [Crossref] [Google Scholar]
- Devi M, Kaur G, Kour A, Kumar A, Singh HP, et al. Metabolic engineering of plant secondary metabolites: Prospects and technological challenges. Front Plant Sci. 2023;14:1171154. [Crossref] [Google Scholar] [PubMed]
- Shankar A, Sharma H, Kumar A, Rani R. Integration of omics tools for metabolic pathway engineering in plants. Biotechnol Adv. 2022;60:108004.
- Wang X, He Y, Zhang Y, Li Y, Tan H, et al. SlMYB12 regulates flavonol synthesis in three cherry tomato varieties. Sci Rep. 2018;8:19214. [Crossref] [Google Scholar] [PubMed]
- Pulido P, Perello C, Rodriguez-Concepcion M. Differential contribution of the first two enzymes of the MEP pathway to carotenoid biosynthesis in plants. Front Plant Sci. 2016;7:1344. [Crossref] [Google Scholar] [PubMed]
- Barja V, Rodríguez-Concepción M. Plant geranylgeranyl diphosphate synthases: every (gene) family has a story. Trends Plant Sci. 2021;26(10):1076-86. [Crossref] [Google Scholar] [PubMed]
- Sazegari S, Niazi A, Shahriari-Ahmadi F, Moshtaghi N, Zare N. CrMYC1 contributes to catharanthine and ajmalicine accumulation in Catharanthus roseus hairy roots. Hortic Environ Biotechnol. 2022;63(5):709–17. [Crossref] [Google Scholar]
- De Geyter N, Gholami A, Goormachtig S, Goossens A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012;17(6):349-59. [Crossref] [Google Scholar] [PubMed]
- Butelli E, Titta L, Giorgio M, Mock HP, Matros A, et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol. 2008;26(11):1301–8. [Crossref] [Google Scholar] [PubMed]
- Sazegari S, Niazi A, Shahriari-Ahmadi F, Moshtaghi N, Zare N. CrMYC1 contributes to catharanthine and ajmalicine accumulation by regulating the TIA pathway in Catharanthus roseus hairy roots. Hortic Environ Biotechnol. 2022;63(5):709–17. [Crossref] [Google Scholar]
- Belchí-Navarro S, Almagro L, Lijavetzky D, Bru R, Pedreño MA. Enhanced extracellular production of trans-resveratrol in Vitis vinifera suspension cultured cells by using cyclodextrins and methyljasmonate. Plant Cell Rep. 2012;31(1):81–9. [Crossref] [Google Scholar] [PubMed]
- Gutiérrez-Carbajal MG, Monforte-González M, Miranda-Ham MDL, Godoy-Hernández G, Vázquez-Flota F. Induction of capsaicinoid synthesis in Capsicum chinense cell cultures by salicylic acid or methyl jasmonate. Biol Plant. 2010;54(3):430–4. [Crossref] [Google Scholar]
- Ochoa-Alejo N. Capsaicin Accumulation in Capsicum spp. Suspension Cultures. Plant Cell Culture Protocols. Totowa, NJ: Humana Press; 2006. p. 327-35. [Crossref] [Google Scholar] [PubMed]
- Bais HP, Walker TS, Schweizer HP, Vivanco JM. Root specific elicitation and antimicrobial activity of rosmarinic acid in hairy root cultures of Ocimum basilicum. Plant Physiol Biochem. 2002;40(11):983–95. [Crossref] [Google Scholar]
- Wang J, Mao S, Liang M, Zhang W, Chen F, et al. Preharvest Methyl Jasmonate Treatment Increased Glucosinolate Biosynthesis, Sulforaphane Accumulation, and Antioxidant Activity of Broccoli. Antioxidants. 2022;11(7):1298. [Crossref] [Google Scholar] [PubMed]
- Nehme A, Awada Z, Kobeissy F, Mazurier F, Zibara K. Coupling Large-Scale Omics data for deciphering systems complexity. RNA Technologies and Their Applications. Cham: Springer; 2018. p. 153–72. [Crossref] [Google Scholar]
- Karahalil B. Overview of Systems biology and Omics Technologies. Curr Med Chem. 2016;23(37):4221–30. [Crossref] [Google Scholar] [PubMed]
- Choi H, Pavelka N. When One and One Gives More than Two: Challenges and Opportunities of Integrative Omics. Front Genet. 2012;2:105. [Crossref] [Google Scholar] [PubMed]
- Yang D, Du X, Yang Z, Liang Z, Guo Z, et, al. Transcriptomics, proteomics, and metabolomics to reveal mechanisms underlying plant secondary metabolism. Eng Life Sci. 2014;14(5):456–66. [Crossref] [Google Scholar]
- Anugraha AC, Thomas T, Thomas TD. Multiomics approach in medicinal plants. Plant Secondary Metabolism Engineering. Academic Press; 2022. p. 589–602. [Crossref] [Google Scholar]
- Seydel C. Diving deeper into the proteome. Nat Methods. 2022;19(9):1036–40. [Crossref] [Google Scholar] [PubMed]
- Zhu F, Wen W, Cheng Y, Alseekh S, Fernie AR. Integrating multiomics data accelerates elucidation of plant primary and secondary metabolic pathways. aBIOTECH. 2023;4(1):47–56. [Crossref] [Google Scholar] [PubMed]
- Hao Y, Zhang Z, Luo E, Yang J, Wang S. Plant metabolomics: applications and challenges in the era of multi-omics big data. aBIOTECH. 2025;6(1):116–32. [Crossref] [Google Scholar] [PubMed]
- Gelder V, Kristen, Lindner N, Hanson DA, Zhou, et, al. Strangers in a foreign land: 'Yeastizing' plant enzymes. Microb Biotechnol. 2024;17(9):e14525. [Crossref] [Google Scholar] [PubMed]
- Ramirez-Estrada K, Vidal-Limon H, Hidalgo D, Moyano E, Golenioswki M, et al. Elicitation, an effective strategy for the biotechnological production of bioactive high-added value compounds in plant cell factories. Molecules. 2016;21(2):182. [Crossref] [Google Scholar] [PubMed]
- Sharma S, Shahzad A. Bioreactors: A Rapid Approach for Secondary Metabolite Production. Production of Biomass and Bioactive Compounds Using Bioreactor Technology. Dordrecht: Springer; 2013. p. 25–49. [Crossref] [Google Scholar]
- Cotten C, Reed JL. Mechanistic analysis of multi-omics datasets to generate kinetic parameters for constraint-based metabolic models. BMC Bioinformatics. 2013;14:32. [Crossref] [Google Scholar] [PubMed]
- Munawar N, Faheem M, Niamat A, Munir A, Khan SH, et al. Regulatory, ethical, social, and biosafety concerns in genome-edited horticultural crops. Genome Editing in Plants. Academic Press; 2024. p. 421–38. [Crossref] [Google Scholar]
- Saifi M, Ashrafi K, Qamar F, Abdin MZ. Regulatory Trends in Engineering Bioactive-Phytocompounds. Plant Sci. 2024;247:112167. [Crossref] [Google Scholar] [PubMed]
- Yang D, Du X, Yang Z, Liang Z, Guo Z, et, al. Transcriptomics, proteomics, and metabolomics to reveal mechanisms underlying plant secondary metabolism. Eng Life Sci. 2014;14(5):456–66. [Crossref] [Google Scholar]
- Nayana BR, Vandana CD, Jayashree V, Sandeep K. Harnessing Machine Learning and Artificial Intelligence for Omics Data Analysis. Artificial Intelligence and Machine Learning in Bioinformatics. New Delhi: AkiNik Publications; 2025. p. 85–94. [Crossref] [Google Scholar]
- Parthiban SV, Vijeesh T, Gayathri T, Shanmugaraj B, Sharma A, et, al. Artificial intelligence-driven systems engineering for next-generation plant-derived biopharmaceuticals. Front Plant Sci. 2023;14:1252166. [Crossref] [Google Scholar] [PubMed]
- Jakočiūnas T, Bonde I, Herrgård MJ, Harrison SJ, Kristensen M, et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–22. [Crossref] [Google Scholar] [PubMed]
- Mitra M, Swain H, Debsarma O, Mandal N. Biotechnological interventions in upscaling of plant secondary metabolites. Biosynthesis of Secondary Metabolites. Burlington, ON: Apple Academic Press; 2023. p. 245–62. [Crossref] [Google Scholar]
- Moses T, Mehrshahi P, Smith AG, Goossens A. Synthetic biology approaches for the production of plant metabolites in unicellular organisms. J Exp Bot. 2017;68(15):4057–74. [Crossref] [Google Scholar] [PubMed]
Article Processing Timeline
| 2-5 Days | Initial Quality & Plagiarism Check |
| 15 Days |
Peer Review Feedback |
| 85% | Acceptance Rate (after peer review) |
| 30-45 Days | Total article processing time |
Indexed In
ResearchBib
Sindexs
OAJI
DOAJ
CrossRef
PubMed
MEDLINE
EBSCO A-Z / Host
OCLC - WorldCat
Journal Flyer